Deubiquitylases (DUBs) are key regulators of the ubiquitin system which cleave ubiquitin moieties from proteins and polyubiquitin chains. Several DUBs have been implicated in various diseases and are attractive drug targets.
We have developed a sensitive and fast assay to quantify in vitro DUB enzyme activity using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Unlike other current assays, this method uses unmodified substrates, such as diubiquitin topoisomers.
Ubiquitylation
Posttranslational modifications with ubiquitin control almost every process in cells. Ubiquitylation is facilitated by ubiquitin-activating (E1s), ubiquitin-conjugating (E2s) and ubiquitin ligase enzymes (E3s). Ubiquitin can be attached to substrate proteins as a single moiety or in the form of polymeric chains in which successive ubiquitin molecules are connected through specific isopeptide bonds. These bonds can be formed on any of the eight primary amines of the ubiquitin molecule and thus can achieve a remarkable complexity, termed the ubiquitin code1, in which the different chain topologies serve distinct signalling functions.
Deubiquitylation
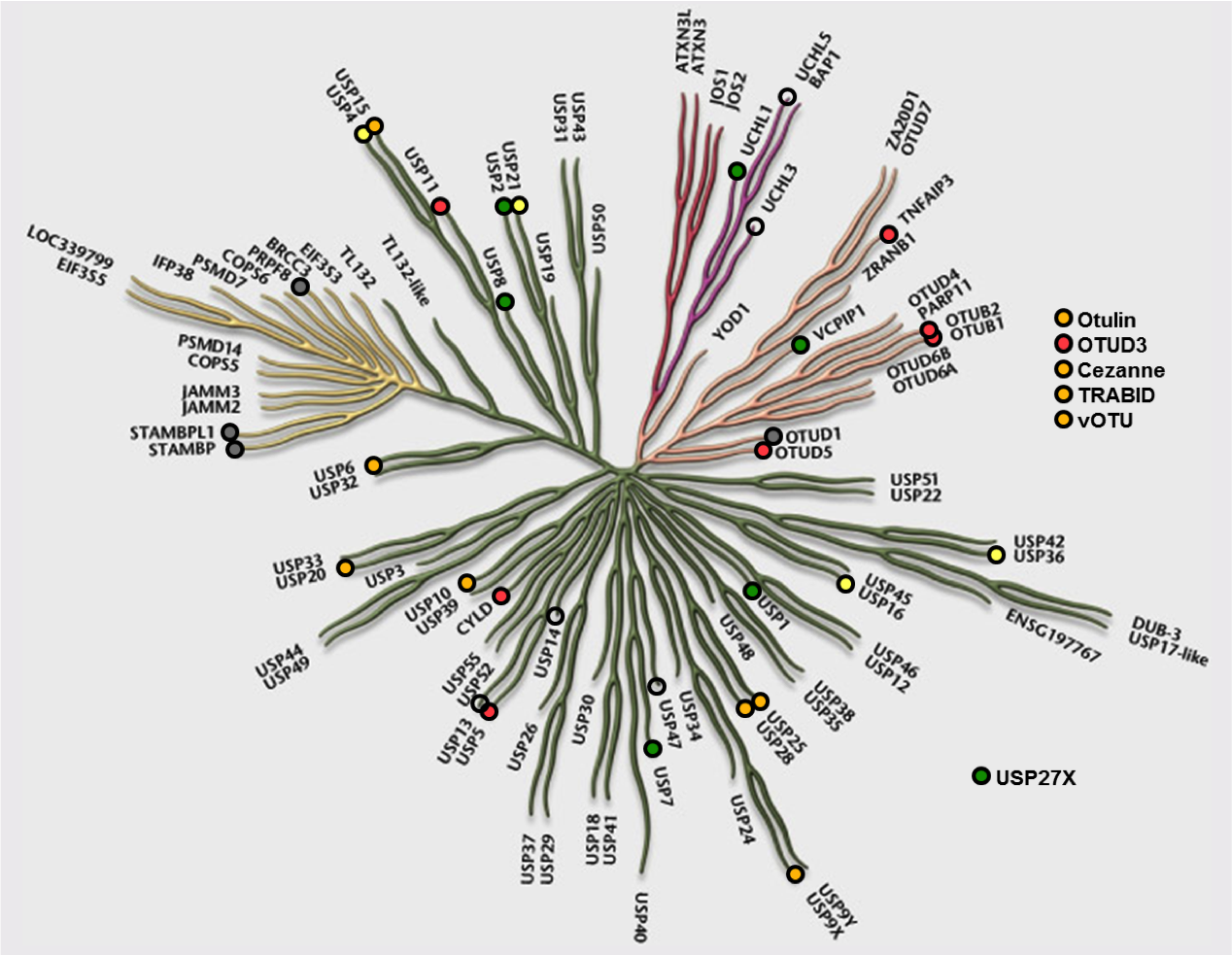
DUBs have an essential role in ubiquitin homeostasis by catalysing the editing and disassembly of polyubiquitin chains. Furthermore, DUBs also perform signalling functions by the regulatory deubiquitylation of target proteins controlling proteasome-dependent protein degradation, endocytosis, DNA repair and kinase activation. Not surprisingly, DUBs have been implicated in a number of diseases such as cancer, inflammation, neurodegeneration/Parkinson’s disease and, due to their potentially drugable active sites, are considered attractive drug targets.
Screening
To screen for DUB inhibitors, current methods make use of non-physiological substrates including linear fusion of ubiquitin to a reporter protein such as phospholipase or yellow fluorescent protein in a Fluorescent Resonance Energy Transfer assay format. Moreover, fusions of fluorogenic reporters such as Rhodamine110 or 7-amino-4-methylcoumarinto the C-terminal glycine of ubiquitin are also widely deployed. However, these substrates are not suitable for assessing the linkage specificity of DUBs. Furthermore, as these are artificial substrates that do not contain physiological isopeptide bonds, screening assays using these substrates could potentially identify compounds that might not inhibit the deubiquitylation of physiological substrates. To circumvent these issues it is possible to undertake DUB assays with more physiologically related diubiquitin molecules.
MALDI-TOF DUB assay.
We have developed a fast and sensitive assay to analyse in vitro activity and specificity of DUBs by MALDI-TOF mass spectro- metry, termed the MALDI-TOF DUB assay. In this assay, we quantitate the amount of monoubiquitin generated by the in vitro cleavage of specific diubiquitin topoisomers by DUBs
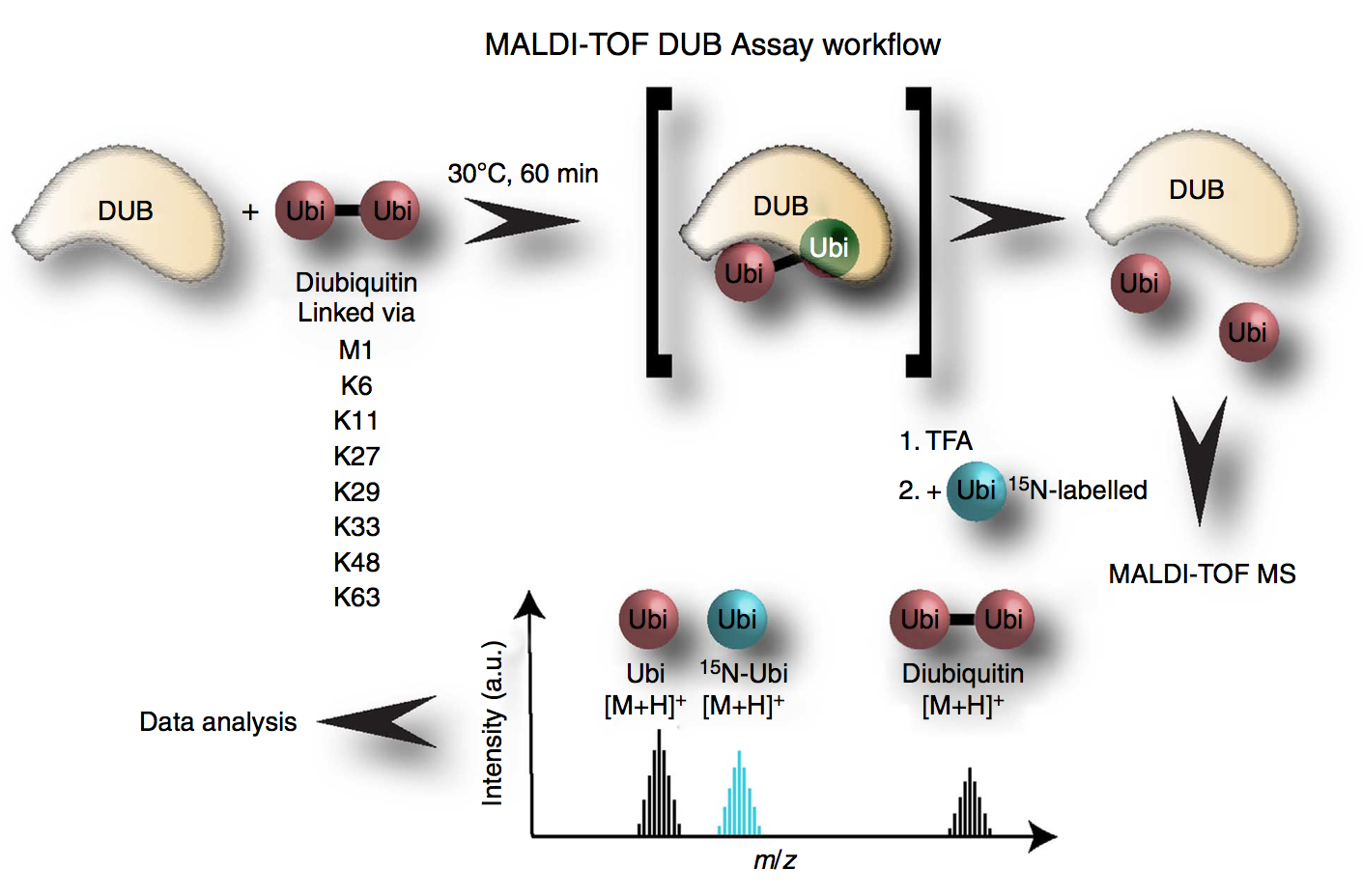
Due to the use of 1,536-sample targets, the assay is suitable for robotic automation and thus high-throughput screening. We circumvented spot-to-spot and shot-to-shot irreproducibility in MALDI ionization by using isotopically labelled ubiquitin as an internal standard as it guarantees identical extraction, crystallization and gas-phase behaviour.
Overall, this setup allowed us to achieve very high precision, accuracy and linearity of measurements over concentrations of almost three orders of magnitude. The advantages compared with the commonly used assays with fluorogenic ubiquitin substrates are the use of substrates which are more physiological and the ability to analyse chain linkage specificity. Moreover, compared with current techniques using SDS–PAGE, our assay is considerably faster (2–4h for the acquisition of 1,536 data points) and more sensitive, thus requiring vastly reduced amounts of diubiquitin substrate. It should be noted that the assay is currently pipetted manually and due to addition of matrix and trifluoroacetic acid (TFA), only 3.3% of the initial reaction mixture is utilized for the mass spectrometry analysis. Thus, after optimization, it should be feasible to scale down reaction amounts at least another 20-fold using nanoliter dispensing robotics representing a nearly 600-fold reduction in amounts of diubiquitin needed in current low-throughput assays.